-
Micropropagation plays an important role in the propagation of numerous ornamental plants[1] as well as many other horticultural crops. In the ornamental aroid Caladium, meristem tip culture can be used to effectively produce clean stock material free of pathogens[1]. Hartman[1] successfully regenerated caladiums using meristem-tip culture that were determined to be free of Dasheen Mosaic Virus (DsMV) using visual observations and indicator plants. The explants were cultured on a modified Murashige & Skoog[2] medium with myo-inositol instead of meso-inositol and edamin was omitted. Growth regulators used were kinetin (1 mg·L−1) and IAA (15 mg·L−1). While Hartman's study[1] did not report any off types in plantlets in vivo, further studies have reported issues with the occurrence of varying phenotypes. Unfortunately, the appearance of somaclonal variants following transplant in vivo has caused caladium producers to be cautious about the use of the technique.
Somaclonal variation was defined by Larkin & Scowcroft[3] to characterize plants derived through cell, tissue, or organ culture that differ from the original plant. With the goal of micropropagation being to produce true-to-type clones, any variation from the original explant can be a major issue for growers and their customers. A significant change or mutation in a single culture can be multiplied to thousands of plants in production causing major losses if the mutation leads to an obvious and/or deleterious phenotypic change. The source of these variations has been attributed to a variety of factors including plant growth regulators, lighting conditions, or exposure to sterilants during explant sterilization[4].
Somaclonal variation has been a common occurrence among Araceae genera with reports of variants in Alocasia, Anthurium, and Syngonium just to name a few[5]. Common plant characteristics include changes in leaf variegation, leaf shape, and plant size. Cultivar selection from somaclonal variants has been shown to have high success in multiple Araceae genera greatly reducing the otherwise long breeding cycles. Somaclonal variants of Dieffenbachia, Aglaonema, Anthurium, and Syngonium have all been selected for cultivar release with unique leaf variegations and flower colors[6−9]. In Dieffenbachia somaclonal variation has been used as an excellent source of variation for cultivar development with reports of somaclonal variation rates of up to 40.4% in 'Camouflage' cultivar[9].
Chu & Yazawa[10] reported the first somaclonal variation in four caladium cultivars and multiple instances have been reported since. Changes in leaf shape and color, vein color, and petiole attachment and color have all been reported[11−13]. Many researchers have performed molecular investigations revealing changes in gene copy numbers, mutations, chromosome number, and epigenetic changes in somaclonal variants of multiple species[14−20]. The age of explant and the concentration and type of plant growth regulators used have been shown to have a large effect on somaclonal variation frequency in caladium. It has been shown that regenerated caladiums in vitro produce the lowest number of somaclonal variants in shoot tips[11]. The number of variants produced increased as explant leaf age increased with expanded leaves producing the most somaclonal variants. A study by Ahmed et al.[12] investigated the effect of type and concentration of auxins on caladium leaf explants. The study found that the percentage of somaclonal variants increased with each increase in auxin concentration and with NAA, IBA, IAA, and 2,4-D, respectively. Certain caladium cultivars have been shown to produce higher frequencies of somaclonal variation than others. Ahmed et al.[21] identified leaf-color-stable and leaf-color-unstable cultivars. Color-unstable cultivars had somaclonal variation rates of 21%–43% and color-stable cultivars had only 0−6% of variants. Thongpukdee et al.[22] reported up to 87.32% of 'Jao Ying' regenerated plants showing phenotypic variation. While these high frequencies of variation negatively impact micropropagation of caladium, they can provide new novel phenotypes for selection by breeding programs.
The production of polyploids has been proven possible in caladiums through the production of 'Tapestry' tetraploids (2n = 60)[16]. In the study, leaf segments were treated with 0.05, 0.01, and 0.2% (w/v) colchicine for 2, 4, and 6 d and were cultured on Murashige and Skoog (MS) media supplemented with 1 mg·L−1 NAA and 3 mg·L−1 BA. The treatment yielded 10 different somaclonal variant types with five of those being identified as tetraploids. The leaf treatment in 0.2% colchicine for 4 d had the highest tetraploid induction rate of 24.14%. The regenerated tetraploids had thicker leaves and petioles attributing to a more upright growth habit potentially advantageous to container performance[16]. A tetraploid disomic (2n = 4x−2 = 58) or double trisomic (2n = 4x−1−1 = 58) 'Red Flash' caladium along with nine other groups of somaclonal variants were regenerated by subjecting leaf explants to in vitro culture[17]. The culture media used for regeneration was composed of MS media with 1 mg·L−1 BA and 0.1 mg·L−1 NAA or 0.1 mg·L−1 2,4-D. Although a number of tetraploids were produced, the presence of aneuploidy in many of the regenerated plants makes their use for plant breeding questionable.
While polyploids developed were overall undesirable as they lacked plant fullness and visually appealing qualities, they did have thicker petioles and leaves than their diploid counterparts. With these stouter features, there is potential for polyploids to have improved resistance to high winds and weather. Recently, Parrish & Deng[23] discovered naturally occurring caladium triploids with potential production value. Like tetraploids that have been produced, they had thicker leaves and petioles in addition to rounder leaves. However, these plants did produce more leaves and had a fuller plant appearance than has been seen with tetraploids. Crossing induced tetraploids with diploids to produce triploids is of high interest with hopes of combining the hardier traits of tetraploids with more visually attractive traits of the commercial diploids.
Although experiments have been conducted examining somaclonal variants derived from meristem tips and from auxin-containing media, little information is available on somaclonal variation from meristem tip cultured caladiums micropropagated on auxin-free media. The goal of this study was to identify polyploid somaclonal variants with clean genome duplications that could be used for plant breeding. The objectives of the present study were to: 1) investigate the suitability of auxin-free micropropagation to induce somaclonal variants in caladium, and 2) reveal and characterize potential morphological, cytological, and molecular changes in somaclonal variants compared to wild-types. Information gained from these analyses might be valuable to plant breeders, nurseries, and caladium growers.
-
Somaclonal variants used in the present study were generated in a previous virus elimination study conducted in Dr. Michael Kane's laboratory in Gainesville, FL, U.S.A. (Kane et al., unpublished data). Tubers from 15 caladium cultivars ('Aaron', 'Brandywine', 'Candidum', 'Carolyn Whorton', 'Cherry Tart', 'Fairytale Princess', 'Freida Hemple', 'Pink Beauty', 'Postman Joyner', 'Red Flash', 'Rosebud', 'Tapestry', 'White Christmas', 'White Queen', and 'White Wing') (Supplemental Table S1) were soaked in Subdue Maxx® (20 µL·L−1) (Syngenta Crop Protection, Greensborough, NC, U.S.A.), a systemic fungicide with the active ingredient mefenoxam. Pre-soaked tubers were then planted in 3.8-L plastic containers filled with sterile perlite. Pots were hand watered daily and shoots were forced from tubers in three weeks.
New caladium shoots were removed and sterilized in 50% ethanol (v/v) for 1 min and then 2.1% sodium hypochlorite (w/v) with two drops of TWEEN®-20 for 10 min. Meristem tips (apical meristematic dome with one or two leaf primordia) approximately 0.2−1 mm long were removed from isolated shoots under a stereoscope and inoculated on the meristem-tip establishment medium. The medium consisted of 4.43 g·L−1 MS basal Salts[2] (Product ID: M530, PhytoTechnology Laboratories, Lenexa, KS, U.S.A.), 148 mg·L−1 of NaH2PO4, 30 g·L−1 sucrose, 0.4 mg·L−1 thiamine, 100 mg·L−1 m-inositol, 0.25 mg·L−1 6-(γ,γ-dimethylallylamino) purine (2ip), and solidified with 2 g·L−1 Gellan Gum CultureGel™ (Product ID: G434, PhytoTechnology Laboratories) (pH = 5.7).
Thirty days after inoculation, caladium cultures were indexed for cultivable bacteria and fungi according to Kane et al.[24] in both liquid and on semi-solid (10 g·L−1 agar) Leifert & Waites Sterility Test Medium (45.22 g·L−1)[25] (Product ID: L476, PhytoTechnology Laboratories). Caladium cultures free of cultivable bacteria and fungi were transferred to a Stage 2 shoot multiplication medium containing 4.43 g·L−1 LS basal salts[26] (Product ID: L689, PhytoTechnology Laboratories), 30 g·L−1 sucrose, 2 mg·L−1 2iP, 148 mg·L−1 of NaH2PO4, and 2 g·L−1 Gellan Gum CultureGel™ (pH = 5.7). Culture conditions for the entirety of in vitro culture were a 16-h photoperiod provided be cool white fluorescent bulbs at 35 μM m−2·s−1 and temperature maintained at 21 ± 2 °C. Plantlets were sub-cultured on fresh multiplication medium every 4−6 weeks (Fig. 1). Over 12,000 caladiums were micropropagated over the 15 cultivars.

Figure 1.
Caladium micropropagation in baby food jars (6 cm wide and 10 cm tall) (left) and during sub-culturing in a large glass Petri dish (approximately 15 cm diameter) (right). During micropropagation, plantlets were sub-cultured to new multiplication media every 4−6 weeks.
For acclimatization of the micropropagated plantlets in the greenhouse, caladium shoots with multiple roots were harvested from the culture jars and rinsed in tap water to remove residual media. The harvested and washed caladiums were then transferred to 48-cell plastic trays (6.05 × 3.81 × 5.72 cm/cell) filled with Pro-Mix® BX general purpose potting media (Premier Tech Horticulture, Quakertown, PA, U.S.A.). A transparent dome was placed on each tray to maintain high humidity and removed daily to hand water. Domes were removed after three weeks, and plants were individually transferred to 3.8-L plastic pots filled with Pro-Mix® BX potting media. After three months growth, apparent somaclonal variant plants were identified from the population visually. The percentage of somaclonal variants based on visual inspection was relatively low (0 to 14.6%), except for 'White Christmas' which recorded an abnormally large incidence of 67.7%. Most abnormalities were leaf color or variegation changes and others had thicker, shiny leaves indicating potential polyploidization. These variants were maintained in pots for at least 1 year prior to evaluation.
Experimental conditions
-
Three somaclonal variants from 'Freida Hemple' (FH-2, FH-3, and FH-4), three variants from 'White Christmas' (WC-2, WC-7, and WC-16), one variant from 'White Wing' (WW-1), and one variant from 'White Queen' (WQ-1) were selected for this study. Variant tubers were harvested in the fall of 2020 and allowed to cure for three months before planting. To use as controls in the experiment, wildtype 'White Christmas,' 'Freida Hemple,' and 'White Wing' plants were forced from tubers gathered from the caladium collection maintained at the University of Florida/IFAS's Gulf Coast Research and Education Center (UF/IFAS/GCREC; Wimauma, FL, U.S.A.). Additionally, wildtype plants of 'White Queen' caladiums were forced from tubers donated by Florida Boys Caladiums (Lake Placid, FL, U.S.A.). All wildtype caladium tubers were grown under field conditions and asexually propagated through tuber division as described by Deng & Harbaugh [27].
All tubers were planted in 3.8-L plastic containers filled with the commercial potting mix Jolly Gardener Pro-Line C/B Growing Mix (Jolly Gardener, Poland, ME, U.S.A.) mixed with 6.51 kg·m–3 of a controlled-release fertilizer (Osmocote, 15N–3.9P–10K, 5- to 6-month release at 21 °C; The Scotts Company, Marysville, OH, U.S.A.). Plants were maintained in a temperature-controlled greenhouse set at 29.4 °C during the day and 21.1 °C at night under natural light. Pots were hand watered every 2−3 d. Pots were arranged in a randomized complete block design (RCBD) with five replicates. The experimental unit was single plants grown in individual 3.8-L plastic containers.
Morphological characterization
-
Leaf characteristics were measured once all plants reached maturity (three or more leaves) at 17 weeks after planting. A stainless-steel ruler was used to measure leaf length and width of mature leaves from the leaf apex to the bottom of the basal nodes and across the widest part of the leaf, respectively. A digital caliper (Fowler & NSK Max-Cal, Japan) was used to capture petiole diameter, leaf blade thickness, and leaf main vein thickness. For each of the five replicates two to three measurements were taken for leaf length, leaf width, petiole diameter, leaf blade thickness, and leaf main vein thickness. To record stomata length, width, and density, nail polish imprints were taken from the abaxial side of three mature leaves per accession. To estimate stomata density, 15 fields of view were examined for each accession in the study. Thirty stomata length and width measurements were taken over three leaves per accession using ImageJ (version 1.53e; National Institutes of Health, Bethesda, MD, U.S.A.)[28].
Nuclear DNA content
-
The protocol recommended by Doležel et al.[29] was used to determine nuclear DNA content. Based on previous caladium DNA content reports, rye [Secale cereal 'Daňkovské' (16.19 pg/2C)] was selected as the appropriate internal standard. Newly unfurled caladium leaves and rye leaf blades were rinsed under tap water and approximately 30 mg of leaf tissue each from the caladium leaf sample and the internal standard were co-chopped in 1 mL of the LBO1 buffer and 50 µL of RNase (Sigma-Aldrich; 1 mg·mL−1) with a sharp razor blade. The nuclei-containing solution was then filtered through a nylon mesh filter (50 µm pore) and 50 µL of the DNA fluorochrome propidium iodide (Sigma-Aldrich; 1 mg·mL−1) was added. The sample tube was then fed into a Cyflow® Ploidy Analyser (Sysmex Europe GmbH, Norderstedt, Germany) flow cytometer. Three flow cytometrical analyses were run per leaf sample and three separate plants were analyzed per accession. The DNA content of each analysis was calculated using the formula recommended by Doležel et al.[29]: nuclear DNA content of caladium samples = nuclear DNA content of 'Daňkovské' rye (16.19 pg/2C) × (mean fluorescence value of caladium samples ÷ mean fluorescence value of the internal standard).
-
Prior to 10:00 AM, rapidly growing caladium root tips (1 cm) were collected from each of the variants and wildtype plants and quickly immersed in 0.002 M 8-hydroxyquinoline solution in the dark at ~24 °C for 2.5 h and then 4 °C for 3.5 h. Root tips were then rinsed with deionized water three times and placed in a fixative solution (3 methanol: 1 acetic acid, v/v) overnight at 4 °C. The fixed roots were rinsed again in deionized water three times before digestion in 1N HCl at room temperature (~24 °C) for 23 min. The digested roots were quickly washed three more times in deionized water before staining overnight in 2% aceto-carmine solution (Carolina Biology Supply Company, Burlington, NC, U.S.A.). The following day, root caps were removed with a sharp scalpel and the meristematic tissue was transferred to a clean glass slide. A drop of 2% aceto-carmine solution was applied and covered with a coverslip. The blunt end of a scalpel was used to tap the coverslip, squashing the cells and spreading out the chromosomes. Cells were viewed using an Olympus BH-2 bright field microscope (Olympus, Center Valley, PA, U.S.A.). Cells with darkly stained chromosomes were photographed under the 100 × objective lens with a Sony DSC-F717 camera (Sony Corporation of America, NY, U.S.A.) fixed to the microscope.
Molecular marker analysis
-
The eight somaclonal variants and four respective wildtypes were analyzed using seven caladium-specific simple sequence repeat (SSR) markers designed by Gong & Deng[30]. A Qiagen DNeasy Plant Mini Kit (Qiagen, Cat. No: 69104) was used to independently extract the genomic DNA from two plants for each of the samples. DNA sample concentrations for each of the 24 samples were estimated using a Nanodrop™ 1000 spectrophotometer (Thermo Scientific, Odessa, TX, U.S.A.). Genomic DNA preps were diluted to a working concentration of 8 ng·µL−1. The SSR primers were synthesized by Eurofins Genomics LLC (Louisville, KY, USA) with an M13 tail (5'-CCCAGTCACGACGTTG-3') attached to the 5' end of each forward primer. Each PCR reaction contained 32 ng of DNA template (8 ng·µL−1), 0.25 units of Taq DNA polymerase, 1× Standard Taq Reaction Buffer, 2 mM dNTPs, 2 pmol of reverse primer, 0.2 pmol of forward primer with M13 tail, and 1.8 pmol of Tide Flour 6 labeled M13 tail primer (New England Biolabs, Ipswich, MA, U.S.A.). An Eppendorf Pro S Mastercycler 6325 (Eppendorf, AG, Hamburg, Germany) was used for PCR amplification running a touchdown program: denaturation at 94 °C for 2 min, seven cycles at 94 °C for 45 s, 68 °C (decrease 2 °C progressively each cycle) for 45 s, and 72 °C for 60 s and then 30 cycles of 45 s at 94 °C, 45 s at 54 °C and 60 s at 72 °C, and a final extension at 72 °C for 5 min. PCR products were diluted 1:10–1:25 depending on primer pair and then denatured at 94 °C for 3 min before loading into gel. PCR products (0.8 µL) were separated on 6.5% polyacrylamide gels and visualized on a LI-COR 4300 DNA analyzer (LI-COR, Lincoln, NE, U.S.A.). Gels were electrophoresed at 1500 V for 1 h and 45 min and the gels were maintained at 45°C for the duration of the electrophoresis.
Statistical analysis
-
To determine significant differences (P ≤ 0.05) among caladium accessions, analysis of variance (ANOVA) was performed on R 4.1.0 (R Core Team, 2021). Tukey's honestly significant difference test was used to separate means when significant differences were found.
-
All somaclonal variants analyzed in this study could easily be distinguished from their wildtype upon visual inspection of leaf coloring. 'White Wing' is identified by its white leaf background with white veins and a green margin, 'Freida Hemple' has a red background with red veins and a green margin, 'White Christmas' has a white leaf background with green veins and a white margin, and 'White Queen' has a white background with red veins and white margin (Fig. 2). 'White Wing' somaclonal variant WW-1 was nearly identical to 'White Wing' but had slightly thicker green margins than the wildtype. 'Freida Hemple' variants were very similar as well in leaf color with each of the variants matching the wild type with red backgrounds, red veins, and green margins. The three variants were differentiated by the thickness of the green margin around the edge of the leaf with FH-3 having the smallest margins followed by FH-2 and FH-4. 'White Christmas' somaclonal variant WC-2 and 'White Christmas' were nearly identical in coloring with a white background and green veins and WC-16 had more prominent green veins. WC-7 had different coloring with a green background with white interveinal segments and green veins. The somaclonal variant of cultivar 'White Queen' WQ-1 was easily differentiated from its wildtype having a green leaf background color with red veins and red patch of color around the petiole attachment.
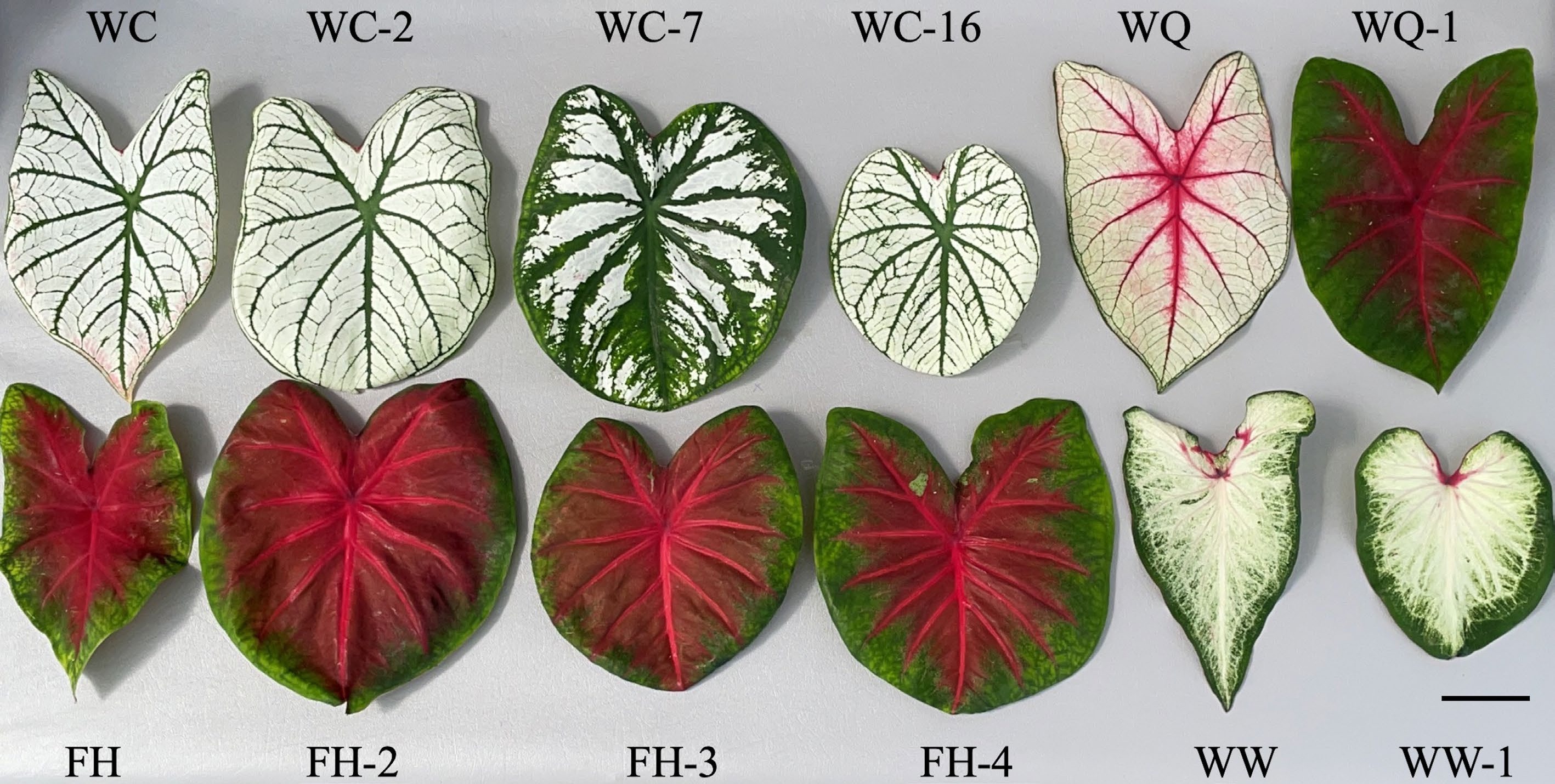
Figure 2.
Typical leaves of caladium somaclonal variants next to their respective wildtypes. Images were taken at time of data collection 17 weeks after planting at the University of Florida/IFAS's Gulf Coast Research and Education Center (U.S.A.). Scale bar (bottom right corner) = 5 cm.
These somaclonal variants also differed from their respective wildtypes in other leaf morphological aspects. WW-1 showed significant changes from 'White Wing' in all areas of leaf morphological measurements recorded. Most notable was a 39% increase in leaf thickness and a 26% increase in petiole thickness (Table 1). 'Freida Hemple' variants also displayed some drastic changes from the wild type, particularly regarding leaf thickness. F47-2, F47-3, and F47-4 all showed approximately 50% increases in leaf thickness. F47-4 also recorded a 79% increase in leaf main vein thickness and 42% increase in petiole diameter. The three somaclonal variants of 'White Christmas' recorded smaller changes in petiole diameter and main vein thickness but did show large increases in leaf thickness. W25-7 had the greatest change in leaf thickness with an increase of 55% when compared to 'White Christmas', with the other two variants recording an approximately 45% increase. Surprisingly, W25-2 and W25-16 recorded 10 and 12% decreases in leaf main vein thickness. The single somaclonal variant of 'White Queen' had smaller changes in leaf morphology with the highest change being 10% increase in petiole diameter. Overall, all variants that were confirmed to be tetraploids (see below) showed an approximately 20% decrease leaf length to width ratios when compared to their respective wild types. A 44.4% increase in leaf thickness was observed among tetraploid variants when compared to diploids. Tetraploid variants also recorded a 30% increase in leaf main vein thickness and petiole diameter.
Table 1. Caladium leaf morphological data comparing somaclonal variants to their respective wildtypes. Data were collected on five plants grown at the University of Florida/IFAS's Gulf Coast Research and Education Center (U.S.A.).
Cultivar/somaclonal
variant (ploidy level)Leaf length/
width ratio
(mean ± SD)Leaf thickness (mm)
(mean ± SD)Leaf main vein
thickness (mm)
(mean ± SD)Petiole diam (mm)
(mean ± SD)Stomata density
(no. /mm2)
(mean ± SD)Stomata length
(µm)
(mean ± SD)Stomata width
(µm)
(mean ± SD)White Wing (2x) 1.72 ± 0.20 aa 0.31 ± 0.02 b 1.34 ± 0.28 b 2.84 ± 0.30 b 55.47 ± 7.64 a 31.03 ± 3.51 b 17.37 ± 2.22 b WW-1 (4x) 1.37 ± 0.16 b 0.43 ± 0.03 a 1.58 ± 0.26 a 3.57 ± 1.65 a 34.53 ± 7.16 b 42.03 ± 4.26 a 21.90 ± 3.09 a Freida Hemple (2x) 1.58 ± 0.13 a 0.26 ± 0.02 b 1.40 ± 0.27 b 3.72 ± 0.34 b 104.17 ± 7.27 a 28.33 ± 3.39 b 16.77 ± 2.43 c FH-2 (4x) 1.27 ± 0.14 b 0.39 ± 0.02 a 1.89 ± 0.33 ab 4.73 ± 0.70 a 38.33 ± 2.93 b 45.60 ± 5.92 a 23.03 ± 3.00 a FH-3 (4x) 1.25 ± 0.11 b 0.39 ± 0.04 a 1.86 ± 0.35 ab 4.80 ± 0.95 a 40.26 ± 3.48 b 47.30 ± 4.72 a 22.10 ± 2.73 a FH-4 (4x) 1.24 ± 0.12 b 0.40 ± 0.02 a 2.51 ± 1.61 a 5.29 ± 0.70 a 40.16 ± 2.50 b 46.20 ± 4.38 a 19.90 ± 2.52 b White Christmas (2x) 1.55 ± 0.14 a 0.25 ± 0.03 c 1.66 ± 0.38 ab 3.56 ± 0.52 b 55.68 ± 7.42 a 30.20 ± 3.89 c 19.23 ± 1.85 b WC-2 (4x) 1.25 ± 0.16 b 0.37 ± 0.03 ab 1.49 ± 0.47 b 3.70 ± 0.77 b 16.56 ± 3.57 c 38.57 ± 4.33 a 22.03 ± 1.92 a WC-7 (4x) 1.25 ± 0.15 b 0.39 ± 0.02 a 1.89 ± 0.39 a 4.44 ± 0.42 a 16.56 ± 4.70 c 44.53 ± 4.76 a 23.53 ± 2.57 a WC-16 (4x) 1.25 ± 0.09 b 0.36 ± 0.03 b 1.47 ± 0.30 b 3.73 ± 0.54 b 28.49 ± 5.69 b 45.50 ± 4.61 b 23.60 ± 2.88 a White Queen (2x) 1.52 ± 0.12 a 0.26 ± 0.01 b 1.31 ± 0.18 a 3.18 ± 0.28 b 71.61 ± 7.08 b 30.57 ± 3.13 a 16.10 ± 1.69 a WQ-1 (2x) 1.41 ± 0.12 b 0.28 ± 0.02 a 1.29 ± 0.13 a 3.50 ± 0.48 a 97.34 ± 7.30 a 28.70 ± 2.39 b 15.47 ± 1.70 a a Means followed by the same letter within each column and group are not significantly different by Tukey's honestly significant difference test at the 5% level of significance. WW-1 had a 38% reduction in stomata density and a 35% and 26% increase in stomata length and width from its wild type (Fig. 3). 'Freida Hemple' somaclonal variants each had a > 60% decrease in stomata density and a > 60% increase in stomata length. The stomata density of W25-2 and W25-7 were identical with 70% fewer stomata than 'White Christmas'. W25-16 had a 49% reduction in stomata density and had a 51% increase in stomata length. While other variants recorded a decrease in stomata density and an increase in size, WQ-1 had the opposite occurrence. The 'White Queen' variant had 36% more stomata and reduction in length and width of 6% and 4%.
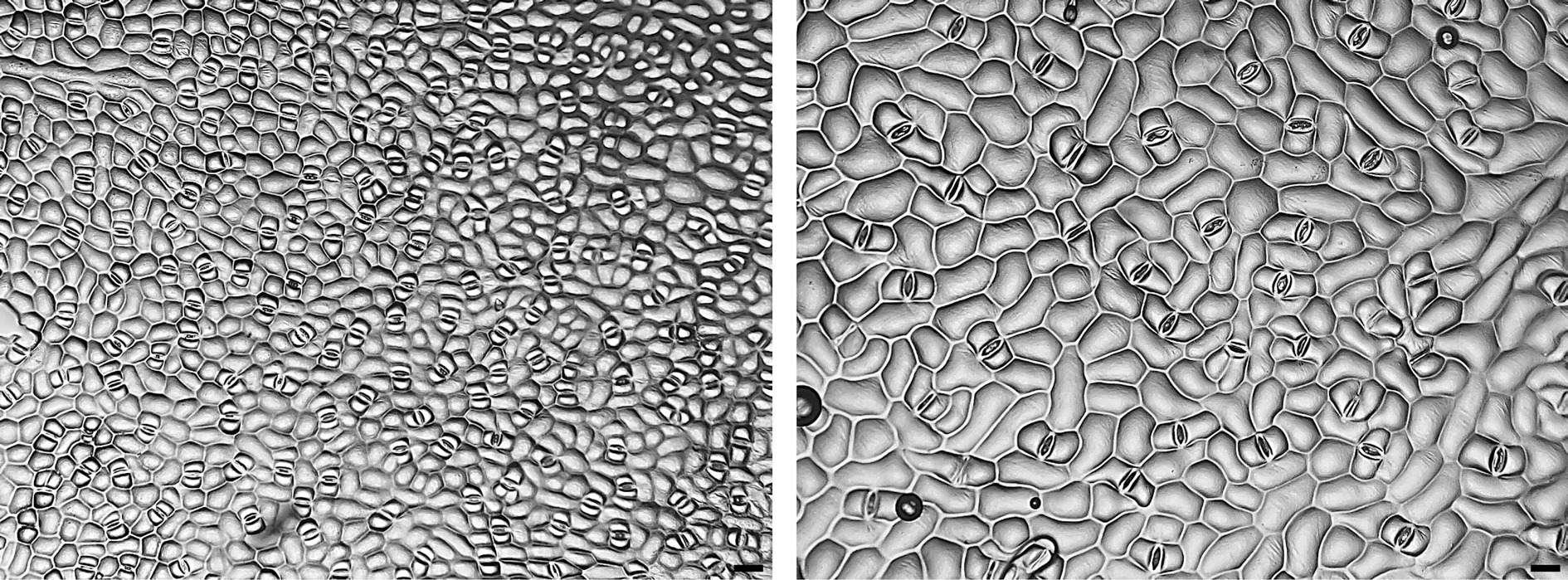
Figure 3.
Micrographs (×100) of stomata imprints from caladiums ‘Freida Hemple’ (diploid wildtype, left) and FH-4 (tetraploid variant, right). Clear nail polish was used to make stomata imprints on the abaxial side of the caladium leaves to be observed under a bright field microscope. Scale bar (bottom right corner) = 50 µm.
Nuclear DNA content
-
Clear changes in nuclear DNA content were detected by the production of sharp histogram peaks using flow cytometry. Variation between individual runs of the same accession was minimal with ≤ 0.03 standard deviation values. Both diploids (2x) and tetraploids (4x) were identified through nuclear DNA content estimations.
The 'White Wing' variant was identified as a tetraploid with a nuclear DNA content of 19.16 pg·2C−1 (Table 2). The variant recorded a 98% increase in DNA content when compared to its wild type. All variants of 'Freida Hemple' were found to be tetraploids with an average nuclear DNA content of 18.60 pg·2C−1. Some variation was present among the somaclonal variants with FH-3 and FH-4 differing from each other by only 0.06 pg, but FH-2 had a large increase over FH-4 of 0.40 pg. Similar to 'Freida Hemple' variants, 'White Christmas' somaclonal variants were all found to be tetraploids. WC-2 and WC-16 had nuclear DNA contents of similar size with 105% increases over 'White Christmas'. WC-7 had a slightly higher nuclear DNA content of 18.89 pg·2C−1. Somaclonal variant WQ-1 had a small drop of 1.6% in nuclear DNA content from its wildtype. Tetraploids in the study had an average nuclear DNA content of 18.76 pg·2C−1 compared to the diploids that recorded an average of 9.37 pg·2C−1.
Table 2. 2C nuclear DNA (pg) and somatic chromosome numbers of somaclonal variants and wildtypes from four caladium cultivars. Three plants per selection were used for nine flow cytometrical analyses.
Cultivar/somaclonal
variantNuclear DNA
content (pg/2C)Nuclear DNA content
change compared
to wildtype (%)Metaphases
observedChromosome number Ploidy level White Wing 9.68 ± 0.02 ba 22 34 2x WW-1 19.16 ± 0.03 a 97.9 7 68 4x Freida Hemple 9.42 ± 0.02 d 3 30 2x FH-2 18.89 ± 0.02 a 100.4 4x FH-3 18.43 ± 0.03 c 95.6 4x FH-4 18.49 ± 0.02 b 96.2 2 60 4x White Christmas 9.15 ± 0.02 c 4 30 2x WC-2 18.71 ± 0.02 b 104.5 4x WC-7 18.89 ± 0.03 a 106.5 2 60 4x WC-16 18.72 ± 0.03 b 104.6 4x White Queen 9.38 ± 0.02 a 7 30 2x WQ-1 9.23 ± 0.01 b -1.6 9 30 2x a Means followed by the same letter within each column are not significantly different by Tukey's honestly significant difference test at the 5% level of significance. Chromosome counting
-
A total of 56 root tip somatic cells were squashed and chromosomes counted to verify ploidy level of the caladium accessions and determine if chromosome numbers would match previous reports (2n = 2x = 30) (Fig. 4). 'White Wing' was found to have 34 somatic chromosomes, four additional chromosomes which may explain the higher nuclear DNA content when compared to other diploids. WW-1, which was identified as a tetraploid using ploidy analysis, was found to contain exactly double the chromosomes of 'White Wing' with 2n = 4x = 68 chromosomes. 'Freida Hemple' had 2n = 2x = 30 chromosomes. FH-4 was selected from among the three somaclonal variants to verify the chromosome number associated with the nuclear DNA content. Chromosome squashes revealed a chromosome number of 2n = 4x = 60 for FH-4. Chromosome squashes of 'White Christmas' showed the expected chromosome number of 2n = 2x = 30. WC-7 was selected for chromosome squashing due to its high nuclear DNA content and revealed 2n = 4x = 60 chromosomes. No difference in chromosome number was found between 'White Queen' and its somaclonal variant WQ-1 with both accessions having 2n = 2x = 30 chromosomes.
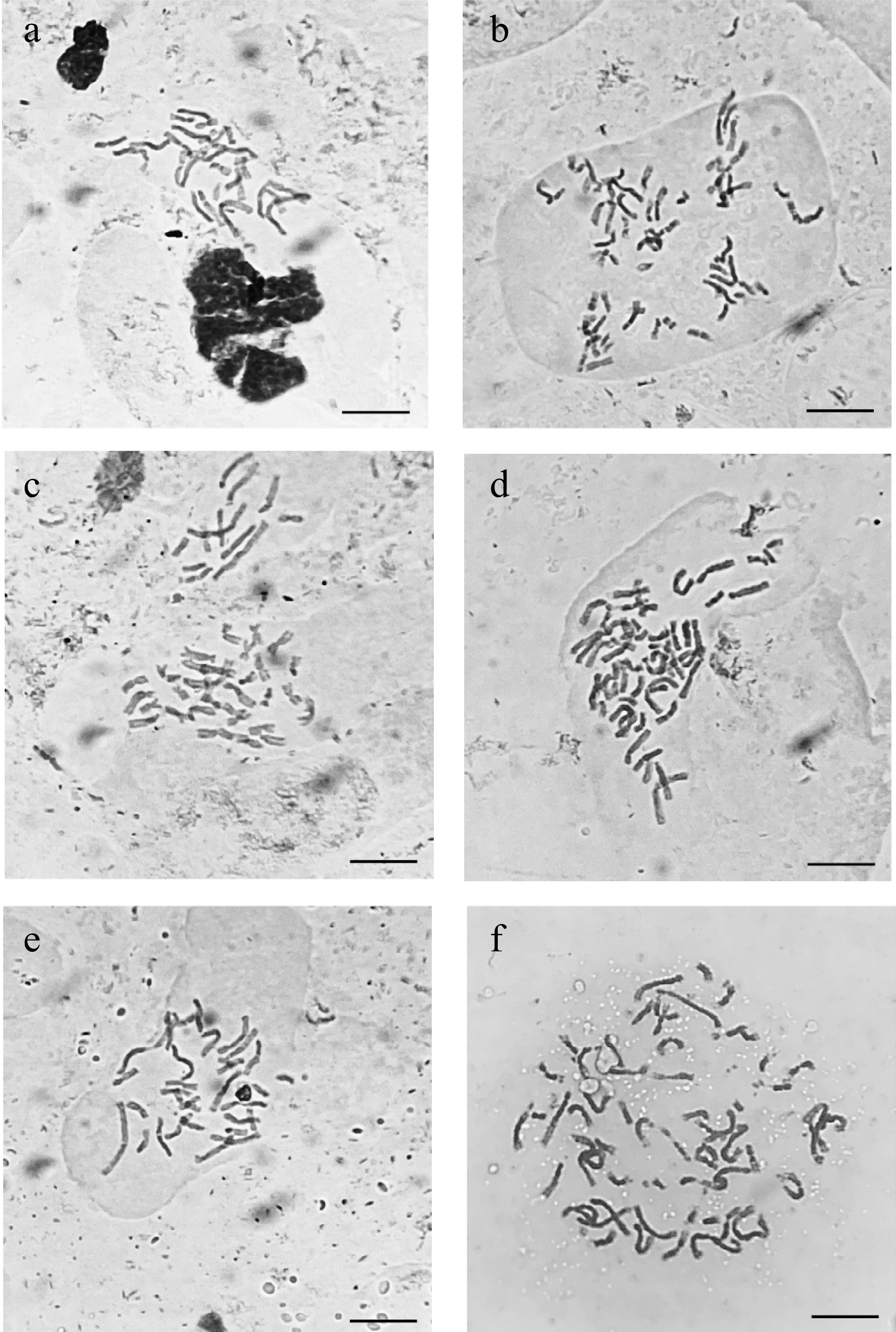
Figure 4.
Micrographs taken under a bright field microscope (×1000) of acetocarmine stained somatic chromosomes from caladium somaclonal variant and wildtype root tips. (a) 'Freida Hemple' (2n = 2x = 30). (b) FH-4 (2n = 4x = 60). (c) 'White Christmas' (2n = 2x = 30). (d) WC-7 (2n = 4x = 60). (e) 'White Wing' (2n = 2x = 30). (f) WW-1 (2n = 4x = 68). Scale bar (bottom right corner) = 10 µm.
SSR marker analysis
-
Out of the seven SSR markers analyzed, only two showed polymorphisms when comparing somaclonal variants to wildtypes. In SSR marker CaM1, 'White Wing' displayed one lighter band on the gel, but WW-1 displayed this band in addition to one much heavier band (Fig. 5, Table 3). Another polymorphism was found in CaM103 where 'White Wing' also displayed one light band and WW-1 displayed the same light band and one heavier band as well. In each of the other 10 accessions, there was consistent banding between somaclonal variants and their respective wildtypes indicating no change in alleles.
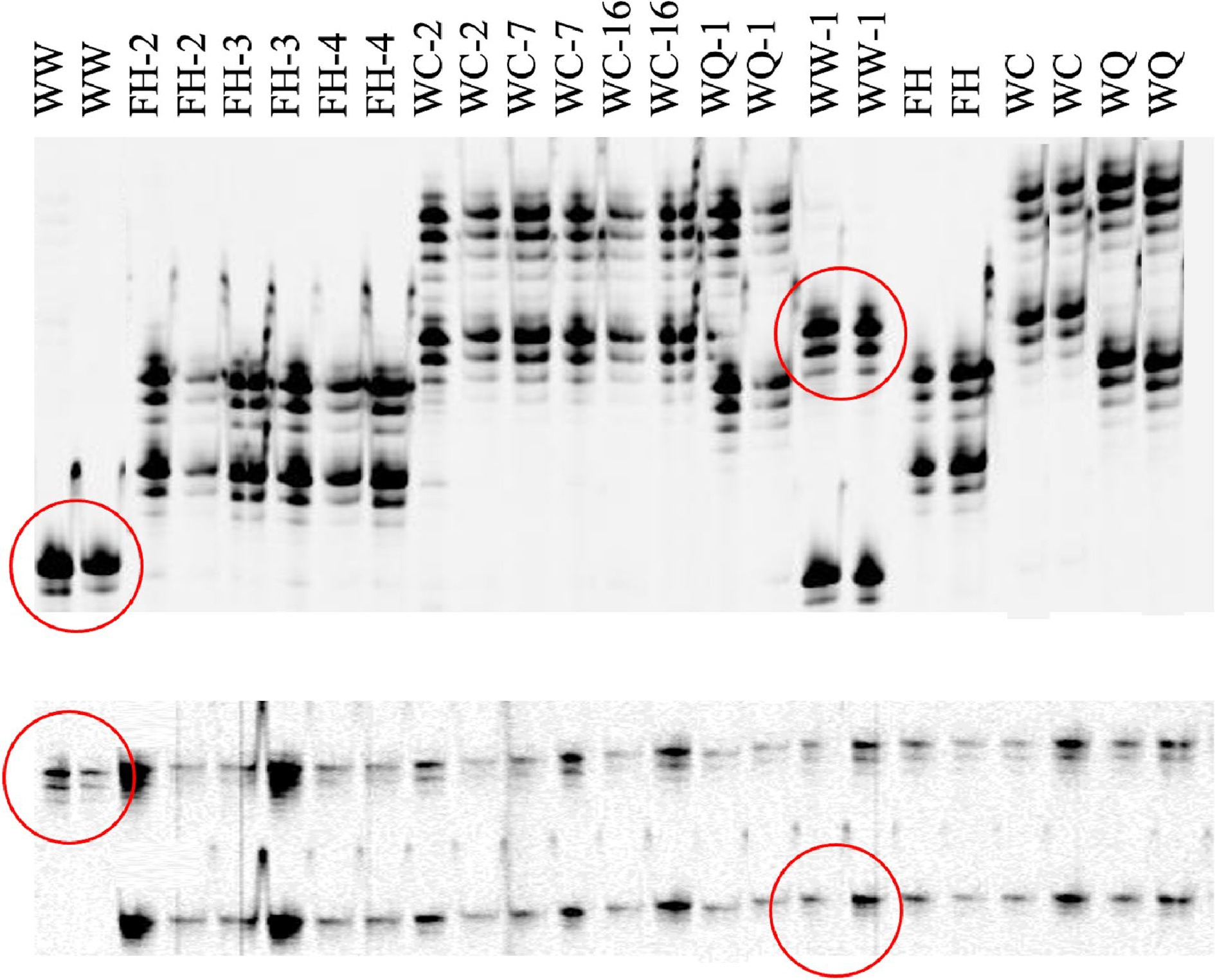
Figure 5.
Polyacrylamide infrared fluorescent gel images generated on a LiCOR 4300 of SSR markers CaM1 (top) and CaM62 (bottom) for caladium somaclonal variants and their wildtypes. Two PCR reactions were conducted using DNA samples from different plants. Wild type caladiums are identified as WW ('White Wing'), FH ('Freida Hemple'), WC (‘White Christmas’), and WQ (‘White Queen’). Polymorphisms between 'White Wing' (diploid wildtype) and WW-1 ('White Wing', tetraploid variant) are outlined in red.
Table 3. Microsatellite marker (SSR) banding patterns from somaclonal variants and their wildtypes. DNA was extracted from two separate plants and used for PCR and electrophoresis. The process was repeated once.
Marker CaM1a CaM18 CaM24 CaM42 CaM48 CaM62 CaM103 Band Numbers 1 2 3 4 5 1 2 1 2 1 2 1 2 3 1 2 1 2 White wing 5 1 2 2 1 2 1 2 1 2 WW-1 2 5 1 2 2 1 2 1 2 1 1 2 Freida hemple 3 4 1 1 2 1 2 1 2 1 2 FH-2 3 4 1 1 2 1 2 1 2 1 2 FH-3 3 4 1 1 2 1 2 1 2 1 2 FH-4 3 4 1 1 2 1 2 1 2 1 2 White christmas 1 2 2 1 2 2 3 1 2 1 2 WC-2 1 2 2 1 2 2 3 1 2 1 2 WC-7 1 2 2 1 2 2 3 1 2 1 2 WC-16 1 2 2 1 2 2 3 1 2 1 2 White Queen 1 3 2 2 2 1 2 1 1 2 WQ-1 1 3 2 2 2 1 2 1 1 2 a Numbers represent the bands produced based on relative size with 1 being the heaviest and n being the smallest. For example, a 1, 2 indicates that a given accession had two bands located on the gel at position 1 and 2 relative to the other bands present for a given marker. -
It was unexpected that the lack of auxins in the tissue culture media would still give rise to multiple tetraploids in caladium. Regenerated caladium cultures from leaf explants on media containing no auxins has reported as low as 6% leaf color somaclonal variants[12]. The control leaf cultures grown on 1 mg·L−1 NAA and 3 mg·L−1 BA in the Cai et al.[16] study were only reported to have one diploid leaf color variant. While no polyploidy was seen in these studies even with the addition of NAA, Ahmed et al.[11] reported many somaclonal variants with round leaves from meristem tip cultures grown on 1 mg·L−1 NAA and 1 mg·L−1 BA. The researchers didn't evaluate ploidy in this study but based on the observations in the current study these plants were likely tetraploids. It is possible that during meristem tip excision and inoculation in vitro, the differentiation of cells from callus could lead to the genome duplication as seen in somaclonal variants within the study. While still present in small amounts, the frequency of somaclonal variants was greatly reduced from previous studies (Parrish, unpublished data).
The 'White Queen' variant WQ-1 interestingly recorded a stomata density close to that of 'Freida Hemple' along with an increase in phenotype similarity. It has been found that leaf background colors (green and lemon) in caladium are controlled by a single locus[31]. It is possible that a single mutation knocked out the white background color of WQ-1 leading to red leaves more similar to 'Freida Hemple'. Overall, the variant has an attractive, vigorous phenotype that could offer advantages over 'Freida Hemple' if it performs well in a production setting.
Tetraploids in this study were found to have a decreased stomata density which is of interest in terms of their resistance to potential plant pathogens. Colletotrichum caladii (Marin et al., Unpublished) is a fungal plant pathogen that has recently become a problem in caladium research plots in central Florida. This fungus primarily enters caladium leaves via the formation of an appressorium[32]. It will be interesting to determine whether an increase in leaf thickness could pose a barrier to the fungal pathogen's infection. Another pathogen having major effects on caladiums is Xanthomonas axonopodis pv. dieffenbachiae, a bacterial pathogen that primarily gains entry to caladiums through their stomata[33]. With a reduction in stomata density, it is possible that caladium tetraploids could possess a barrier to infection for this pathogen. Disease screenings for both of these pathogens are needed to assess any resistance that might be offered by polyploid caladiums.
Tetraploids also had a nearly 1 mm increase in petiole diameter when compared to diploids. This finding is consistent with 'Tapestry' caladium tetraploids where tetraploids recorded a similar increase in petiole thickness[16]. Chen et al.[34] reported 'stronger' petioles in Anthurium andraeanum tetraploids in vitro and in vivo in addition to thicker flower peduncles and longer lasting flowers. Especially during field production in Florida, caladiums often face high winds from tropical cyclones which can cause petiole breakage similar to lodging in many row crops. Thicker petioles could offer resistance for these caladiums to wind damage and thus improve tuber yields in field conditions.
The minimal changes in SSR marker banding patterns support our hypothesis that by not adding auxins to the tissue culture media there would be a reduction in the number of chromosome changes in micropropagated plantlets. Cao et al.[17] demonstrated that SSR markers CaM1 and CaM103 were located on two different chromosomes due to the loss of alleles in one marker but not the other when a chromosome was lost. With knowledge that these two markers are on separate chromosomes, we can conclude that the two alleles gained were separate occurrences. Both CaM1 and CaM103 markers contain an 'AG' motif with 18 and 28 repeats, respectively. It is possible that DNA polymerase slippage could be the culprit of the allele changes between WW-1 and the wildtype 'White Wing'at both loci. Due to the high number of repeats at these microsatellite loci, the nascent DNA strand may have looped out during DNA replication causing an extension of the repeat sequence[35]. If this extension occurred prior to or during genome duplication, the new allele would be present in two of the four chromosomes. It is possible that the cellular event that resulted in endomitosis allowed these slippage events to get past DNA mismatch repair systems.
The ability to produce tetraploid caladiums using an auxin-free media is a testament to the genome duplication efficiency during micropropagation. SSR markers indicate that small chromosomal changes may be occurring due to the absence of auxins in the tissue culture media and therefore could produce near complete chromosome copies during genome duplication events. While tetraploids produced do not possess the ornamental value needed for commercial production, the possibility to use these plants as parents in crosses to produce triploids is intriguing with implications in other aroids. Parrish & Deng[23] discovered two naturally occurring caladium triploid genotypes with increased ornamental value over tetraploids. Plant breeders can use tissue culture as a tool to produce tetraploid somaclonal variants of their elite breeding lines and open the door for triploid production. At the same time breeders will also be able to take advantage of new phenotypes that arise through somaclonal variants and discover new cultivars or breeding materials.
This study was funded in part by the United States Department of Agriculture (USDA) Agricultural Marketing Service (AMS) and Florida Department of Agriculture and Consumer Services (FDACS) Specialty Crop Block Grant Program (FDACS Contract 024844) and USDA hatch projects FLA-GCC-005507 and FLA-GCC-006190.
-
The authors declare that they have no conflict of interest.
- Supplemental Table S1 Main ornamental traits of 15 caladium cultivars used in the study.
- Copyright: © 2023 by the author(s). Published by Maximum Academic Press, Fayetteville, GA. This article is an open access article distributed under Creative Commons Attribution License (CC BY 4.0), visit https://creativecommons.org/licenses/by/4.0/.
-
About this article
Cite this article
Parrish SB, Kane ME, Deng Z. 2023. Morphological, cytogenetic and molecular characterization of new somaclonal variants in four caladium (Caladium × hortulanum) cultivars. Ornamental Plant Research 3:1 doi: 10.48130/OPR-2023-0001
Morphological, cytogenetic and molecular characterization of new somaclonal variants in four caladium (Caladium × hortulanum) cultivars
- Received Date: 11 October 2022
- Accepted Date: 08 December 2022
- Published Online: 17 January 2023
Abstract: The discovery of triploids in caladium has raised strong interest in their use in cultivar development. For triploid production, tetraploids must first be generated from elite breeding material without the introduction of deleterious alleles like has been seen with chemical agents like colchicine and 2,4-D. To meet this new demand, virus eradicated, meristem-tip caladium cultures were grown on auxin-free media and used for generation and evaluation of new somaclonal variants. Over 12,000 micropropagated caladiums were visually screened ex vitro for sports varying from the wild-type phenotype. Eight variants from four cultivars were discovered and characterized for morphological, cytological, and molecular changes. Genome duplication occurred in seven of the variants with an average nuclear DNA content of 18.76 pg·2C−1. The tetraploid variant of cultivar ‘White Wing’ had eight more chromosomes (2n = 4x = 68) than the other tetraploids and exactly double that of its wildtype. All other diploids had the expected 2n = 2x = 30 chromosomes and tetraploids recorded 2n = 4x = 60. Tetraploids had 19% rounder leaves and 44% thicker leaves than the diploids in the study. Tetraploids also had a 49% and 31% increase in stomata length and width, respectively. Molecular marker analysis only revealed two polymorphisms with both occurring in the 'White Wing' variant. Few changes in molecular marker banding could indicate that the tetraploids discovered have a clean duplication of the genome and might effectively be used for new triploid cultivar development.
-
Key words:
- Caladium /
- Ornamental aroid /
- Bulbous plant /
- Somaclonal variant /
- Genome duplication /
- Tetraploid /
- Chromosome doubling /
- Molecular marker /
- Ploidy manipulation













